x = 1
y <- 46 Using R and the tidyverse
R is a programming language, much like Python. It is widely used by data scientists, those using digital humanities techniques, and in the social sciences.
It has some advantages for a novice to programming. Its widely-used platform (called an Integrated Programming Environment- an IDE) for the language called R-Studio makes it relatively easy for beginners. A lot of this is because of developers who have extended the functionality of the base language greatly, particularly a suite of extra functions known collectively as the ‘tidyverse’.
The methods section of this book may be challenging if you are a complete beginner to R and have not learned any other programming language. If you’d like to get started with R and the tidyverse in earnest, the canonical texts is R for Data Science, written by Hadley Wickham.
Getting started
The only requirements to get through these tutorials are to install R and R-Studio, as well as some data which needs to be downloaded separately.
Download R and R-Studio
R and R-Studio are two separate things. R will work without R-studio, but not the other way around, and so it should be downloaded first. Go to the download page, select a download mirror, and download the correct version for your operating system. Follow the installation instructions if you get stuck.
Next, download R-Studio. You’ll want the desktop version and again, download the correct version and follow the instructions. When both of these are installed, open R-Studio, and it should run the underlying software R automatically.
At this point, I would highly recommend reading a beginners guide to R and R-studio, such as this one, to familiarise yourself with the layout and some of the basic functionality of R-Studio. Once you understand where to type and save code, where your files and dataframes live, and how to import data from spreadsheets, you should be good to start experimenting with newspaper data.
R relies on lots of additional packages for full functionality, and you’ll need to install these by using the function install.packages(), followed by the package name, in inverted commas. I recommend doing this to install the tidyverse suite of packages by running install.packages('tidyverse') in the Console window (the bottom-left of the screen in R-Studio) as you’ll end up using this all the time.
Using R
‘Base’ R.
Commands using R without needing any additional packages are often called ‘base’ R. Here are some important ones to know:
You can assign a value to an object using = or <-:
Entering the name of a variable in the console and pressing return will return that value in the console. The same will happen if you enter it in a notebook cell (like here below), and run the cell. This is also true of any R object, such as a dataframe, vector, or list.
y[1] 4You can do basic calculations with +, -, * and /:
x = 1+1
y = 4 - 2
z = x * y
z[1] 4You can compare numbers or variables using == (equals), > (greater than), <, (less than) != (not equal to). These return either TRUE or FALSE:
1 == 1[1] TRUEx > y[1] FALSEx != z[1] TRUEBasic R data structures
It is worth understanding the main types of data that you’ll come across, in your environment window.
A variable is a piece of data stored with a name, which can then be used for various purposes. The simplest of these are single elements, such as a number:
x = 1
x[1] 1Next is a vector. A vector is a list of elements. A vector is created with the command c(), with each item in the vector placed between the brackets, and followed by a comma. If your vector is a vector of words, the words need to be in inverted commas or quotation marks.
fruit = c("apples", "bananas", "oranges", "apples")
colour = c("green", "yellow", "orange", "red")
amount = c(2,5,10,8)Next are dataframes. These are the spreadsheet-like objects, with rows and columns, which you’ll use in most analyses.
You can create a dataframe using the data.frame() command. You just need to pass the function each of your vectors, which will become your columns.
We can also use the glimpse() or str() commands to view some basic information on the dataframe (particularly useful with longer data).
fruit_data = data.frame(fruit, colour, amount, stringsAsFactors = FALSE)
str(fruit_data)'data.frame': 4 obs. of 3 variables:
$ fruit : chr "apples" "bananas" "oranges" "apples"
$ colour: chr "green" "yellow" "orange" "red"
$ amount: num 2 5 10 8Data types
Notice that to the right of the third column, the amount, has <dbl>under it, whereas the other two have
- character
- numeric (real or decimal)
- integer
- logical
- complex
- Raw
The most commonly-used ones you’ll come across are character, numeric, and logical. logical is data which is either TRUE or FALSE. In R, all the items in a vector are coerced to the same type. So if you try to make a vector with a combination of numbers and strings, the numbers will be converted to strings, as in the example below:
fruit = c("apples", 5, "oranges", 3)
str(fruit) chr [1:4] "apples" "5" "oranges" "3"Installing and loading packages:
R is extended through the use of ‘packages’: pre-made sets of functions, usually with a particular task or theme in mind. To work with networks, for example, we’ll use a set of third-party packages. If you complete the exercises using the CSC cloud notebooks, these are already installed for you in most cases. To install a package, use the command install.packages(), and include the package name within quotation marks:
install.packages('igraph')To load a package, use the command library(). This time, the package name is not within quotation marks
library(igraph)Warning: package 'igraph' was built under R version 4.0.5
Attaching package: 'igraph'The following objects are masked from 'package:stats':
decompose, spectrumThe following object is masked from 'package:base':
unionTidyverse
Most of the work in these notebooks is done using a set of packages developed for R called the ‘tidyverse’. These enhance and improve a large range of R functions, with a more intuitive nicer syntax. It’s really a bunch of individual packages for sorting, filtering and plotting data frames. They can be divided into a number of different categories.
All these functions work in the same way. The first argument is the thing you want to operate on. This is nearly always a data frame. After come other arguments, which are often specific columns, or certain variables you want to do something with.
library(tidyverse)Here are a couple of the most important ones
select(), pull()
select() allows you to select columns. You can use names or numbers to pick the columns, and you can use a - sign to select everything but a given column.
Using the fruit data frame we created above: We can select just the fruit and colour columns:
select(fruit_data, fruit, colour) fruit colour
1 apples green
2 bananas yellow
3 oranges orange
4 apples redSelect everything but the colour column:
select(fruit_data, -colour) fruit amount
1 apples 2
2 bananas 5
3 oranges 10
4 apples 8Select the first two columns:
select(fruit_data, 1:2) fruit colour
1 apples green
2 bananas yellow
3 oranges orange
4 apples redgroup_by(), tally(), summarise()
The next group of functions group things together and count them. Sounds boring but you would be amazed by how much of data science just seems to be doing those two things in various combinations.
group_by() puts rows with the same value in a column of your dataframe into a group. Once they’re in a group, you can count them or summarise them by another variable.
First you need to create a new dataframe with the grouped fruit.
grouped_fruit = group_by(fruit_data, fruit)Next we use tally(). This counts all the instances of each fruit group.
tally(grouped_fruit)# A tibble: 3 × 2
fruit n
<chr> <int>
1 apples 2
2 bananas 1
3 oranges 1See? Now the apples are grouped together rather than being two separate rows, and there’s a new column called n, which contains the result of the count.
If we specify that we want to count by something else, we can add that in as a ‘weight’, by adding wt = as an argument in the function.
tally(grouped_fruit, wt = amount)# A tibble: 3 × 2
fruit n
<chr> <dbl>
1 apples 10
2 bananas 5
3 oranges 10That counts the amounts of each fruit, ignoring the colour.
filter()
Another quite obviously useful function. This filters the dataframe based on a condition which you set within the function. The first argument is the data to be filtered. The second is a condition (or multiple condition). The function will return every row where that condition is true.
Just red fruit:
filter(fruit_data, colour == 'red') fruit colour amount
1 apples red 8Just fruit with at least 5 pieces:
filter(fruit_data, amount >=5) fruit colour amount
1 bananas yellow 5
2 oranges orange 10
3 apples red 8You can also filter with multiple terms by using a vector (as above), and the special command %in%:
filter(fruit_data, colour %in% c('red', 'green')) fruit colour amount
1 apples green 2
2 apples red 8slice_max(), slice_min()
These functions return the top or bottom number of rows, ordered by the data in a particular column.
fruit_data %>% slice_max(order_by = amount, n = 1) fruit colour amount
1 oranges orange 10fruit_data %>% slice_min(order_by = amount, n = 1) fruit colour amount
1 apples green 2These can also be used with group_by(), to give the top rows for each group:
fruit_data %>% group_by(fruit) %>% slice_max(order_by = amount, n = 1)# A tibble: 3 × 3
# Groups: fruit [3]
fruit colour amount
<chr> <chr> <dbl>
1 apples red 8
2 bananas yellow 5
3 oranges orange 10Notice it has kept only one row per fruit type, meaning it has kept only the apple row with the highest amount?
sort(), arrange()
Another useful set of functions, often you want to sort things. The function arrange() does this very nicely. You specify the data frame, and the variable you would like to sort by.
arrange(fruit_data, amount) fruit colour amount
1 apples green 2
2 bananas yellow 5
3 apples red 8
4 oranges orange 10Sorting is ascending by default, but you can specify descending using desc():
arrange(fruit_data, desc(amount)) fruit colour amount
1 oranges orange 10
2 apples red 8
3 bananas yellow 5
4 apples green 2If you `sortarrange() by a list of characters, you’ll get alphabetical order:
arrange(fruit_data, fruit) fruit colour amount
1 apples green 2
2 apples red 8
3 bananas yellow 5
4 oranges orange 10You can sort by multiple things:
arrange(fruit_data, fruit, desc(amount)) fruit colour amount
1 apples red 8
2 apples green 2
3 bananas yellow 5
4 oranges orange 10Notice that now red apples are first.
left_join(), inner_join(), anti_join()
Another set of commands we’ll use quite often in this course are the join() ‘family’. Joins are a very powerful but simple way of selecting certain subsets of data, and adding information from multiple tables together.
Let’s make a second table of information giving the delivery day for each fruit type:
fruit_type = c('apples', 'bananas','oranges')
weekday = c('Monday', 'Wednesday', 'Friday')
fruit_days = data.frame(fruit_type, weekday, stringsAsFactors = FALSE)
fruit_days fruit_type weekday
1 apples Monday
2 bananas Wednesday
3 oranges FridayThis can be ‘joined’ to the fruit information, to add the new data on the delivery day, without having to edit the original table (or repeat the information for apples twice). This is done using left_join.
Joins need a common key, a column which allows the join to match the data tables up. It’s important that these are unique (a person’s name makes a bad key by itself, for example, because it’s likely more than one person will share the same name). Usually, we use codes as the join keys. If the columns containing the join keys have different names (as ours do), specify them using the syntax below:
joined_fruit = left_join(fruit_data, fruit_days, by = c("fruit" = "fruit_type"))
joined_fruit fruit colour amount weekday
1 apples green 2 Monday
2 bananas yellow 5 Wednesday
3 oranges orange 10 Friday
4 apples red 8 MondayIn this new dataframe, the correct weekday is now listed beside the relevant fruit type.
Piping
Another useful feature of the tidyverse is that you can ‘pipe’ commands through a bunch of functions, making it easier to follow the logical order of the code. This means that you can do one operation, and pass the result to another operation. The previous dataframe is passed as the first argument of the next function by using the pipe %>% command. It works like this:
fruit_data %>%
filter(colour != 'yellow') %>% # remove any yellow colour fruit
group_by(fruit) %>% # group the fruit by type
tally(amount) %>% # count each group
arrange(desc(n)) # arrange in descending order of the count# A tibble: 2 × 2
fruit n
<chr> <dbl>
1 apples 10
2 oranges 10That code block, written in prose: “take fruit data, remove any yellow colour fruit, count the fruits by type and amount, and arrange in descending order of the total”
Plotting using ggplot()
The tidyverse includes a plotting library called ggplot2. To use it, first use the function ggplot() and specify the dataset you wish to graph using data =. Next, add what is known as a ‘geom’: a function which tells the package to represent the data using a particular geometric form (such as a bar, or a line). These functions begin with the standard form geom_.
Within this geom, you’ll add ‘aesthetics’, which specify to the package which part of the data needs to be mapped to which particular element of the geom. The most common ones include x and y for the x and y axes, color or fill to map colors in your plot to particular data.
ggplot is an advanced package with many options and extensions, which cannot be covered here.
Some examples using the fruit data:
Bar chart of different types of fruit (one each of bananas and oranges, two types of apple)
ggplot(data = fruit_data) + geom_col(aes(x = fruit, y = amount))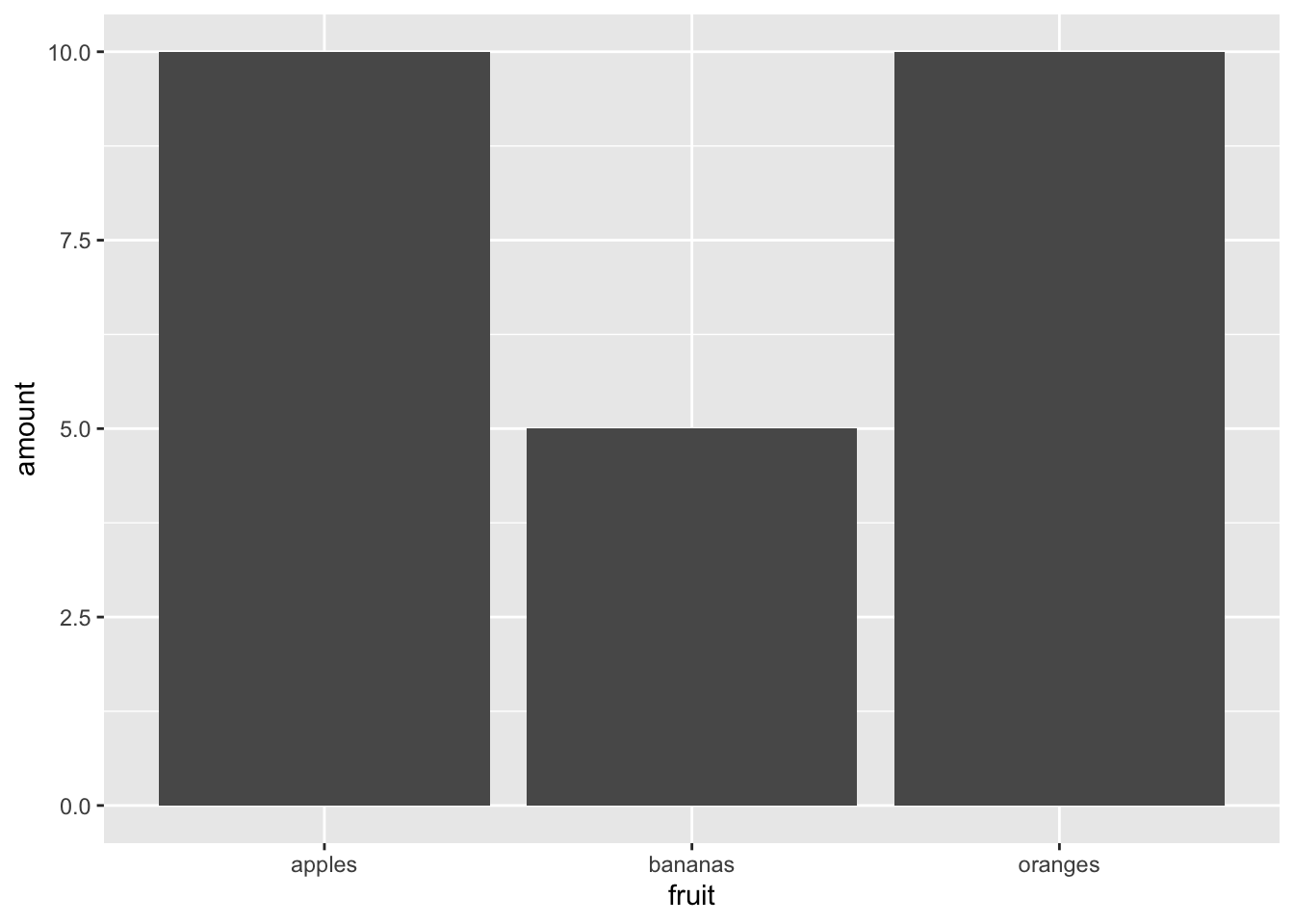
Counting the total amount of fruit:
ggplot(fruit_data) + geom_col(aes(x = fruit, y = amount))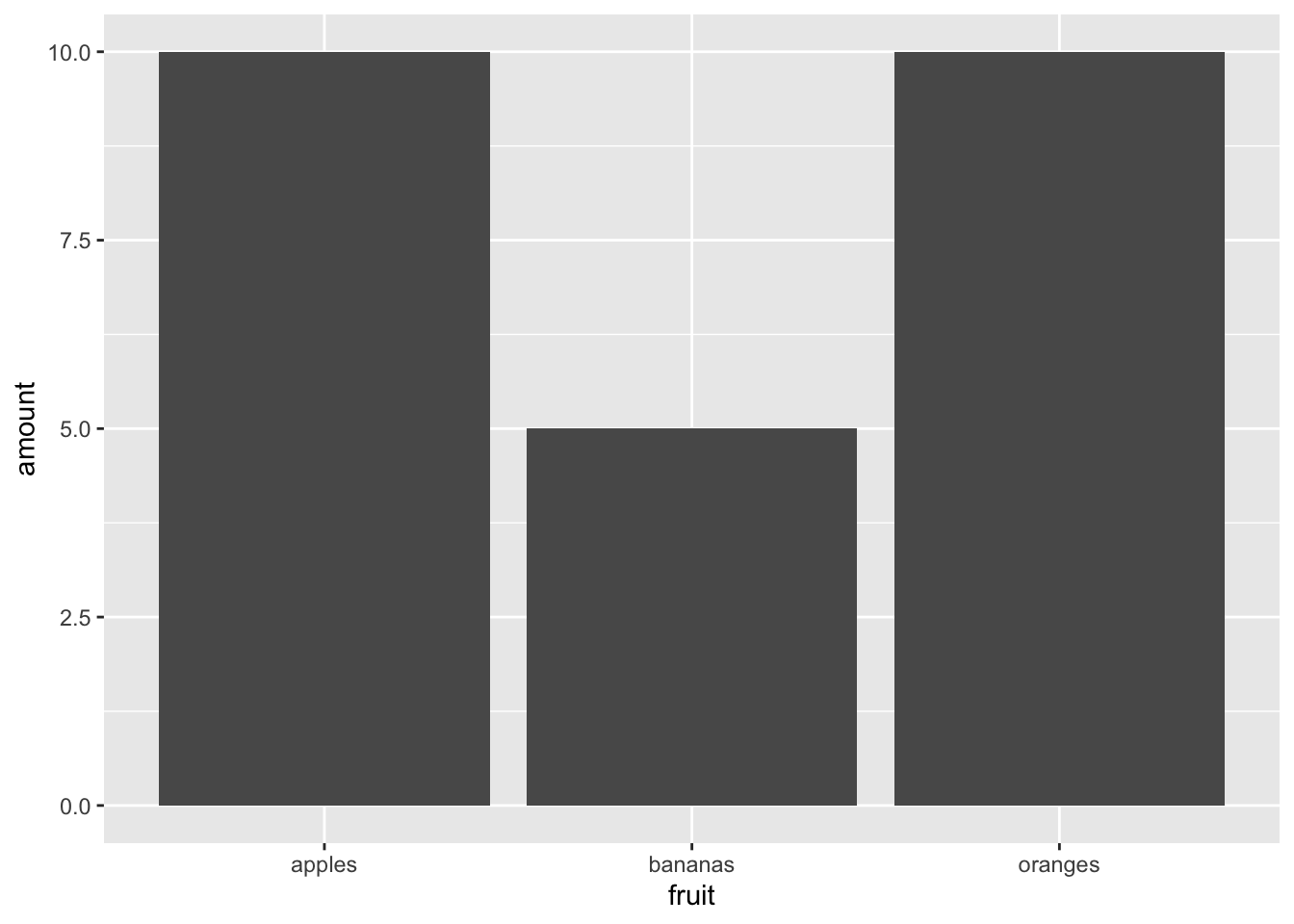
Charting amounts and fruit colours:
ggplot(data = fruit_data) + geom_bar(aes(x = fruit, weight = amount, fill = colour)) 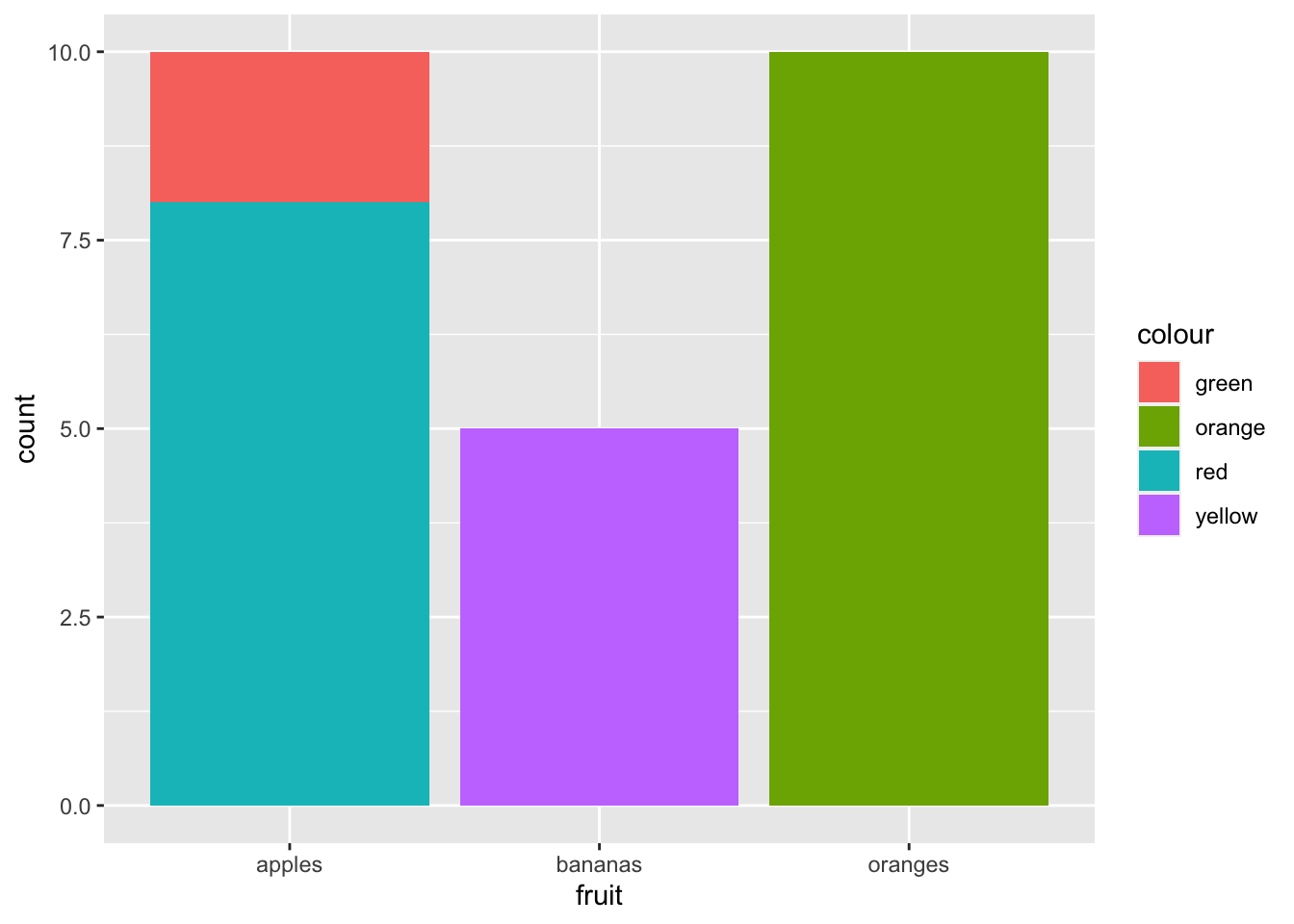
Reading in external data
Most of the time, you’ll be working with external data sources. These most commonly come in the form of comma separated values (.csv) or tab separated values (.tsv). The tidyverse commands to read these are read_csv() and read_tsv. You can also use read_delim(), and specify the type of delimited using delim = ',' or delim = '/t. The path to the file is given as a string to the argument file=.
df = read_csv(file = 'aus_titles.csv') # Read a .csv file as a network, specify the path to the file here.Rows: 2137 Columns: 7
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (4): newspaper_title, state, place_id, place
dbl (3): title_id, latitude, longitude
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.df# A tibble: 2,137 × 7
title_id newspaper_title state place_id place latitude longitude
<dbl> <chr> <chr> <chr> <chr> <dbl> <dbl>
1 984 Adelaide Chronicle and Sout… SA SA00558… Adel… -34.9 139.
2 986 Adelaide Chronicle and Sout… SA SA00558… Adel… -34.9 139.
3 174 Adelaide Morning Chronicle … SA SA00558… Adel… -34.9 139.
4 821 Adelaide Observer (SA : 184… SA SA00558… Adel… -34.9 139.
5 1100 Adelaide Times (SA : 1848 -… SA SA00558… Adel… -34.9 139.
6 277 Adelaider Deutsche Zeitung … SA SA00558… Adel… -34.9 139.
7 434 Adelong and Tumut Express a… NSW NSW81112 Adel… -35.3 148.
8 434 Adelong and Tumut Express a… NSW NSW60433 Tumb… -35.8 148.
9 434 Adelong and Tumut Express a… NSW NSW79906 Tumut -35.3 148.
10 625 Adelong and Tumut Express (… NSW NSW81112 Adel… -35.3 148.
# ℹ 2,127 more rowsNotice that each column has a data type beside it, either
Doing this with newspaper data
Let’s load a dataset of metadata for all the titles held by the library, and practise some counting and sorting on real-world data.
Download from here: British Library Research Repository
You would need to extract into your project folder first, if you’re following along:
read_csv reads the csv from file.
title_list = read_csv('data/BritishAndIrishNewspapersTitleList_20191118.csv')Rows: 24927 Columns: 24
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (18): publication_title, edition, preceding_titles, succeeding_titles, p...
dbl (6): title_id, nid, nlp, first_date_held, publication_date_one, publica...
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.Select some particularly relevant columns:
title_list %>%
select(publication_title,
first_date_held,
last_date_held,
country_of_publication)# A tibble: 24,927 × 4
publication_title first_date_held last_date_held country_of_publication
<chr> <dbl> <chr> <chr>
1 "Corante, or, Newes fr… 1621 1621 The Netherlands
2 "Corante, or, Newes fr… 1621 1621 The Netherlands
3 "Corante, or, Newes fr… 1621 1621 The Netherlands
4 "Corante, or, Newes fr… 1621 1621 England
5 "Courant Newes out of … 1621 1621 The Netherlands
6 "A Relation of the lat… 1622 1622 England
7 "A Relation of the lat… 1622 1622 England
8 "A Relation of the lat… 1622 1622 England
9 "A Relation of the lat… 1622 1622 England
10 "A Relation of the lat… 1622 1622 England
# ℹ 24,917 more rowsArrange in order of the latest date of publication, and then by the first date of publication:
title_list %>%
select(publication_title,
first_date_held,
last_date_held,
country_of_publication) %>%
arrange(desc(last_date_held), first_date_held)# A tibble: 24,927 × 4
publication_title first_date_held last_date_held country_of_publication
<chr> <dbl> <chr> <chr>
1 Shrewsbury chronicle 1773 Continuing England
2 London times|The Times… 1788 Continuing England
3 Observer (London)|Obse… 1791 Continuing England
4 Limerick chronicle 1800 Continuing Ireland
5 Hampshire chronicle|Th… 1816 Continuing England
6 The Inverness Courier,… 1817 Continuing Scotland
7 Sunday times (London)|… 1822 Continuing England
8 The Impartial Reporter… 1825 Continuing Northern Ireland
9 Impartial reporter and… 1825 Continuing Northern Ireland
10 Aberdeen observer 1829 Continuing Scotland
# ℹ 24,917 more rowsGroup and count by country of publication:
title_list %>%
select(publication_title,
first_date_held,
last_date_held,
country_of_publication) %>%
arrange(desc(last_date_held)) %>%
group_by(country_of_publication) %>%
tally()# A tibble: 40 × 2
country_of_publication n
<chr> <int>
1 Bermuda Islands 24
2 Cayman Islands 1
3 England 20465
4 England|Hong Kong 1
5 England|India 2
6 England|Iran 2
7 England|Ireland 10
8 England|Ireland|Northern Ireland 10
9 England|Jamaica 7
10 England|Malta 2
# ℹ 30 more rowsArrange again, this time in descending order of number of titles for each country:
title_list %>%
select(publication_title,
first_date_held,
last_date_held,
country_of_publication) %>%
arrange(desc(last_date_held)) %>%
group_by(country_of_publication) %>%
tally() %>%
arrange(desc(n))# A tibble: 40 × 2
country_of_publication n
<chr> <int>
1 England 20465
2 Scotland 1778
3 Ireland 1050
4 Wales 1019
5 Northern Ireland 415
6 England|Wales 58
7 Bermuda Islands 24
8 England|Scotland 13
9 England|Ireland 10
10 England|Ireland|Northern Ireland 10
# ℹ 30 more rowsFilter only those with more than 100 titles:
title_list %>%
select(publication_title,
first_date_held,
last_date_held,
country_of_publication) %>%
arrange(desc(last_date_held)) %>%
group_by(country_of_publication) %>%
tally() %>%
arrange(desc(n)) %>%
filter(n>=100)# A tibble: 5 × 2
country_of_publication n
<chr> <int>
1 England 20465
2 Scotland 1778
3 Ireland 1050
4 Wales 1019
5 Northern Ireland 415Make a simple bar chart:
title_list %>%
select(publication_title,
first_date_held,
last_date_held,
country_of_publication) %>%
arrange(desc(last_date_held)) %>%
group_by(country_of_publication) %>%
tally() %>%
arrange(desc(n)) %>%
filter(n>=100) %>%
ggplot() +
geom_bar(aes(x = country_of_publication, weight = n))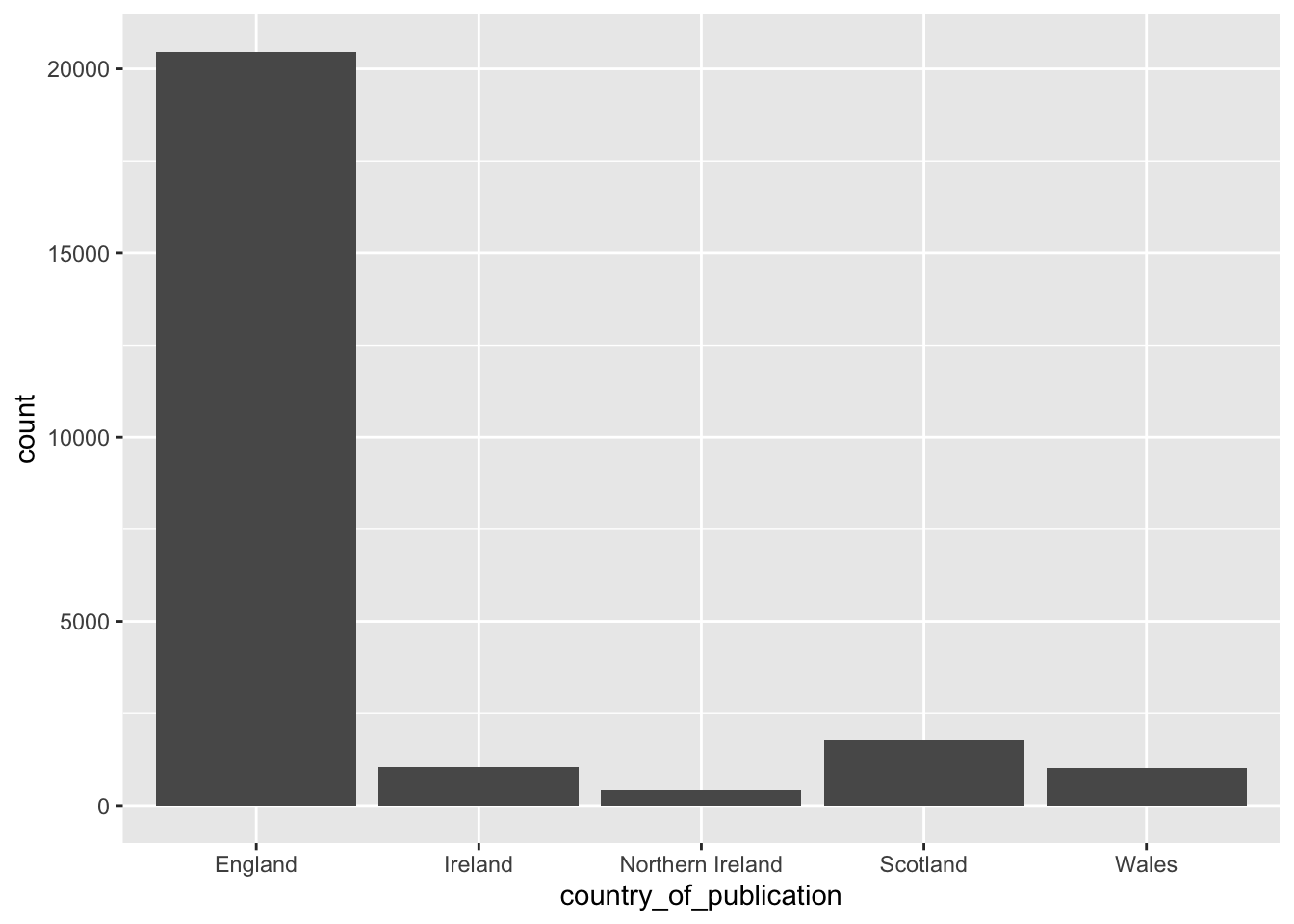
Recommended Reading
This has been a very quick introduction to R. There are lots of resources available to learn more, including:
The Pirate’s Guide to R, a good beginners guide to base R